Burgers equation on a sequence of meshes¶
This demo shows how to solve the Firedrake Burgers equation demo on a sequence of meshes using Goalie. The PDE
is solved on two meshes of the unit square, \(\Omega = [0, 1]^2\). The forward solution is initialised as a sine wave and is nonlinearly advected to the right hand side. See the Firedrake demo for details on the discretisation used.
from firedrake import *
from goalie import *
In this problem, we have a single prognostic variable, \(\mathbf u\). Its name is recorded in a list of field names.
field_names = ["u"]
For each such field, we need to be able to specify how to
build a FunctionSpace, given some mesh. Since there
could be more than one field, function spaces are given as a
dictionary, indexed by the prognostic solution field names.
def get_function_spaces(mesh):
return {"u": VectorFunctionSpace(mesh, "CG", 2)}
In order to solve PDEs using the finite element method, we
require a weak form. For this, Goalie requires a function
that maps the MeshSeq index and a dictionary of
solution data to the form. The form should be
returned inside its own dictionary, with an entry for each equation
being solved for.
The solution Functions are automatically built on the function spaces given
by the get_function_spaces() function and are accessed via the fields
attribute of the MeshSeq. This attribute provides a dictionary of tuples
containing the current and lagged solutions for each field.
Similarly, timestepping information associated with a given subinterval
can be accessed via the TimePartition attribute of
the MeshSeq. For technical reasons, we need to create a Function
in the ‘R’ space (of real numbers) to hold constants.
def get_form(mesh_seq):
def form(index):
# Get the current and lagged solutions
u, u_ = mesh_seq.fields["u"]
# Define constants
R = FunctionSpace(mesh_seq[index], "R", 0)
dt = Function(R).assign(mesh_seq.time_partition.timesteps[index])
nu = Function(R).assign(0.0001)
# Setup variational problem
v = TestFunction(u.function_space())
F = (
inner((u - u_) / dt, v) * dx
+ inner(dot(u, nabla_grad(u)), v) * dx
+ nu * inner(grad(u), grad(v)) * dx
)
return {"u": F}
return form
We have a weak form. The dictionary usage may seem cumbersome when applied to such a simple problem, but it comes in handy when solving adjoint problems associated with coupled systems of equations.
In order to solve the PDE, we need to choose
a time integration routine and solver parameters for the underlying
linear and nonlinear systems. This is achieved below by using a function
solver() whose input is the MeshSeq index. As noted above, the solution
Functions are automatically initialised and accessed via the
fields attribute of the MeshSeq. The lagged solution is assigned
the initial condition for the current subinterval index. For the \(0^{th}\) index,
this will be provided by the initial conditions, otherwise it will be transferred
from the previous mesh in the sequence.
The function should return a generator that yields the solution at each timestep, so that Goalie can efficiently track the solution history. This is done by using the yield statement before progressing to the next timestep.
def get_solver(mesh_seq):
def solver(index):
# Get the current and lagged solutions
u, u_ = mesh_seq.fields["u"]
# Define form
F = mesh_seq.form(index)["u"]
# Time integrate from t_start to t_end
tp = mesh_seq.time_partition
t_start, t_end = tp.subintervals[index]
dt = tp.timesteps[index]
t = t_start
while t < t_end - 1.0e-05:
solve(F == 0, u)
yield
u_.assign(u)
t += dt
return solver
Goalie also requires a function for generating an initial condition from the function space defined on the \(0^{th}\) mesh.
def get_initial_condition(mesh_seq):
fs = mesh_seq.function_spaces["u"][0]
x, y = SpatialCoordinate(mesh_seq[0])
return {"u": assemble(interpolate(as_vector([sin(pi * x), 0]), fs))}
Now that we have the above functions defined, we move onto the concrete parts of the solver. To begin with, we require a sequence of meshes, simulation end time and a timestep.
n = 32
meshes = [
UnitSquareMesh(n, n),
UnitSquareMesh(n, n),
]
end_time = 0.5
dt = 1 / n
These can be used to create a TimePartition for the
problem with two subintervals.
num_subintervals = len(meshes)
time_partition = TimePartition(
end_time,
num_subintervals,
dt,
field_names,
num_timesteps_per_export=2,
)
Finally, we are able to construct a MeshSeq and
solve Burgers equation over the meshes in sequence.
mesh_seq = MeshSeq(
time_partition,
meshes,
get_function_spaces=get_function_spaces,
get_initial_condition=get_initial_condition,
get_form=get_form,
get_solver=get_solver,
)
solutions = mesh_seq.solve_forward()
During the solve_forward() call, the solver that was provided
is applied on the first subinterval. The forward solution at the end
of that subinterval is transferred to the mesh associated with the
second subinterval and used as an initial condition for the same solver
applied again there. Goalie uses a conservative interpolation
operator to transfer solution data between the two meshes. In this
example, the meshes (and hence function spaces) are identical so the
projection operation will in fact be the identity.
The output is a nested dictionary of solution data, indexed by
solution type ("forward" or "forward_old") and then field name
(here "u"). The contents of the inner dictionaries are lists
containing lists of solution Functions, indexed first by
subinterval and then by timestep. That is,
solutions["forward"]["u"][i][j] contains the forward solution
associated with field "u" at the j-th timestep of
subinterval i. Similarly, solutions["forward_old"]["u"][i][j]
contains the forward solution from the timestep prior.
For the purposes of this demo, we plot the solution at each exported
timestep using the plotting driver function plot_snapshots().
fig, axes, tcs = plot_snapshots(
solutions, time_partition, "u", "forward", levels=np.linspace(0, 1, 9)
)
fig.savefig("burgers.jpg")
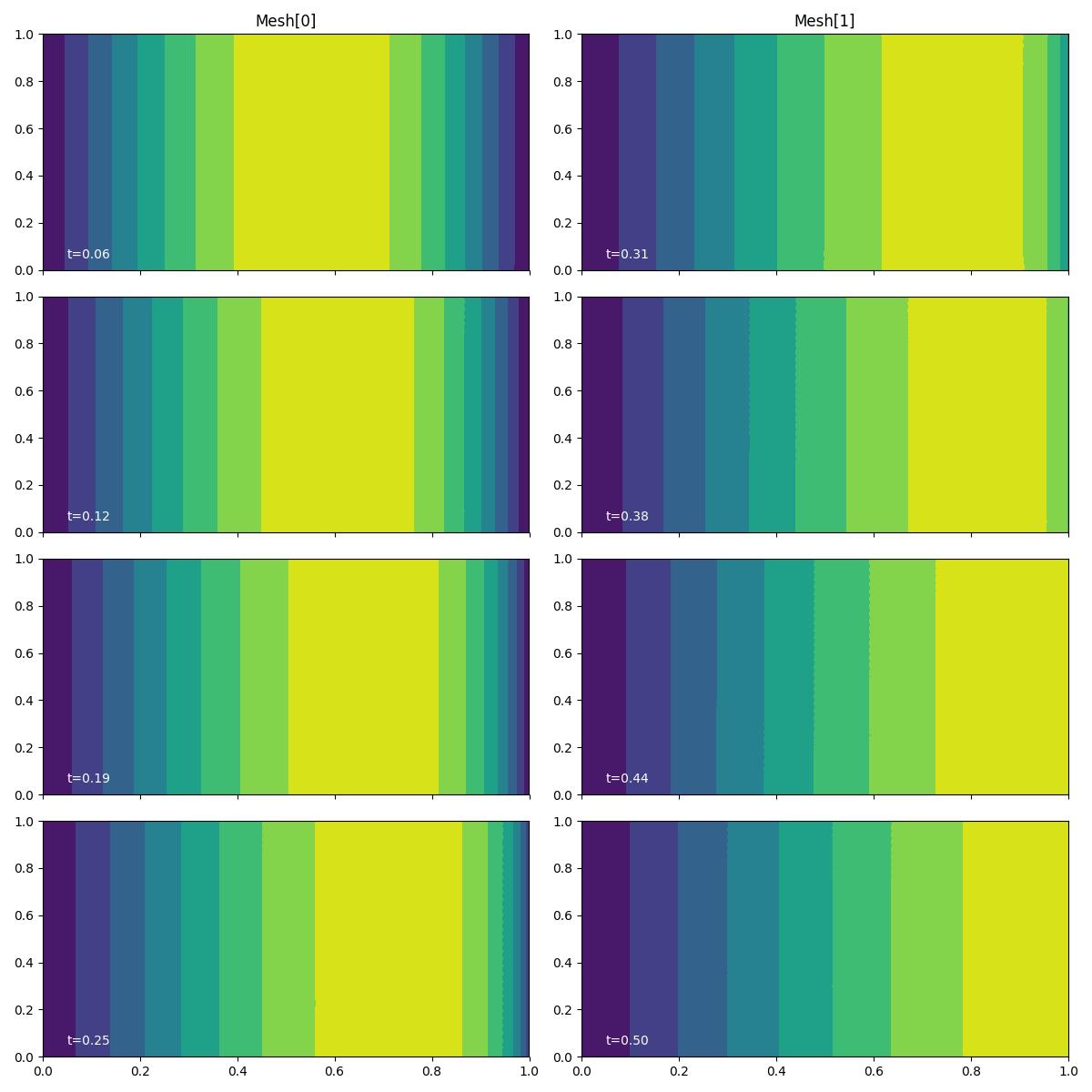
We see that the initial sinusoid is nonlinearly advected Eastwards.
In the next demo, we use Goalie to automatically solve the adjoint problem associated with Burgers equation.
This demo can also be accessed as a Python script.