Using mesh movement to optimise the mesh for PDE solution¶
In this demo we will demonstrate how we might use mesh movement to obtain a mesh that is optimised for solving a particular PDE. The general idea is that we want to reduce numerical error by increasing resolution where the local error is (expected to be) high and decrease it elsewhere.
As an example we will look at the discretisation of the Helmholtz equation
For an explanation of how we can use Firedrake to implement a Finite Element Method (FEM) discretisation of this PDE see the corresponding Firedrake demo. The only changes we introduce is that we choose a different, slightly more interesting solution \(u\) and rhs \(f\)
where \((x_0, y_0)\) is the centre of the Gaussian with width \(w\). Note that here we first choose the solution \(u\) after which we can compute what rhs \(f\) should be, by substitution in the PDE, in order for \(u\) to be the analytical solution. This so-called Method of Manufactured Solutions approach is an easy way to construct PDE configurations for which we know the analytical solution, e.g. for testing purposes.
Based on the code in the Firedrake demo, we first solve the PDE on a uniform mesh. Because our chosen solution does not satisfy homogeneous Neumann boundary conditions, we instead apply Dirichlet boundary conditions based on the chosen analytical solution.
from firedrake import *
from movement import MongeAmpereMover
n = 20
mesh = UnitSquareMesh(n, n) # initial mesh
def u_exact(mesh):
"""Return UFL expression of exact solution"""
x, y = SpatialCoordinate(mesh)
# some arbitrarily chosen centre (x0, y0) and width w
w = Constant(0.1)
x0 = Constant(0.51)
y0 = Constant(0.65)
return exp(-((x - x0) ** 2 + (y - y0) ** 2) / w**2)
def solve_helmholtz(mesh):
"""Solve the Helmholtz PDE on the given mesh"""
V = FunctionSpace(mesh, "CG", 1)
u = TrialFunction(V)
v = TestFunction(V)
u_exact_expr = u_exact(mesh)
f = -div(grad(u_exact_expr)) + u_exact_expr
a = (inner(grad(u), grad(v)) + inner(u, v)) * dx
L = inner(f, v) * dx
u = Function(V)
bcs = DirichletBC(V, u_exact_expr, "on_boundary")
solve(a == L, u, bcs=bcs)
return u
u_h = solve_helmholtz(mesh)
import matplotlib.pyplot as plt
from firedrake.pyplot import tripcolor
fig, axes = plt.subplots()
contours = tripcolor(u_h, axes=axes)
fig.colorbar(contours)
plt.savefig("monge_ampere_helmholtz-initial_solution.jpg")
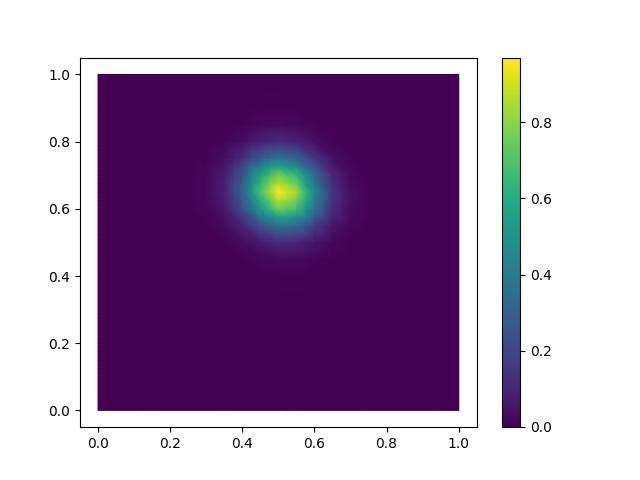
As in the Helmholtz demo, we can compute the L2-norm of the numerical error:
error = u_h - u_exact(mesh)
print("L2-norm error on initial mesh:", sqrt(assemble(dot(error, error) * dx)))
L2-norm error on initial mesh: 0.010233816824277465
We will now try to use mesh movement to optimise the mesh to reduce
this numerical error. We use the same monitor function as
in the previous Monge-Ampère demo
based on the norm of the Hessian of the solution.
In the following implementation we use the exact solution \(u_{\text{exact}}\)
which we have as a symbolic UFL expression, and thus we can also obtain the Hessian
symbolically as grad(grad(u_exact)). To compute its maximum norm however we
do interpolate it to a P1 function space V and take the maximum of the array of
nodal values.
alpha = Constant(5.0)
def monitor_exact(mesh):
V = FunctionSpace(mesh, "CG", 1)
Hnorm = Function(V, name="Hnorm")
H = grad(grad(u_exact(mesh)))
Hnorm.interpolate(sqrt(inner(H, H)))
Hnorm_max = Hnorm.dat.data.max()
m = 1 + alpha * Hnorm / Hnorm_max
return m
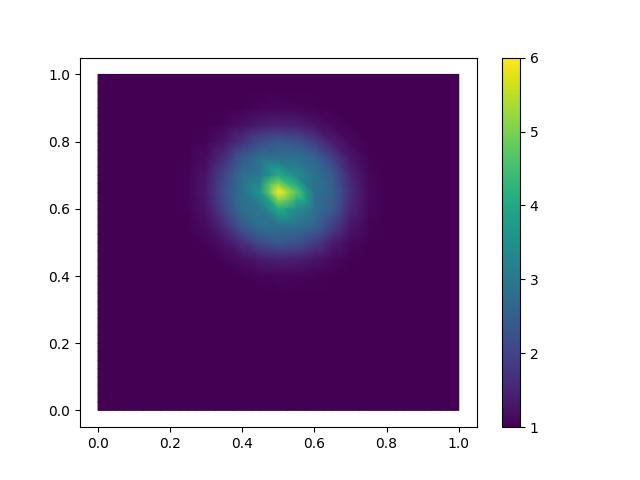
Plot the monitor function on the original mesh:
fig, axes = plt.subplots()
m = Function(u_h, name="monitor")
m.interpolate(monitor_exact(mesh))
contours = tripcolor(m, axes=axes)
fig.colorbar(contours)
plt.savefig("monge_ampere_helmholtz-monitor.jpg")
fig, axes = plt.subplots()
Now we can construct a MongeAmpereMover instance to
adapt the mesh based on this monitor function. We will also time how long the mesh
movement step takes to complete [1].
import time
rtol = 1.0e-08
mover = MongeAmpereMover(mesh, monitor_exact, method="quasi_newton", rtol=rtol)
t0 = time.time()
mover.move()
print(f"Time taken: {time.time() - t0:.2f} seconds")
For every iteration the MongeAmpereMover prints the minimum to maximum ratio of the cell areas in the mesh, the residual in the Monge-Ampère equation, and the coefficient of variation of the cell areas:
0 Volume ratio 4.93 Variation (σ/μ) 4.73e-01 Residual 4.77e-01
1 Volume ratio 2.64 Variation (σ/μ) 2.44e-01 Residual 2.41e-01
2 Volume ratio 1.67 Variation (σ/μ) 1.31e-01 Residual 1.24e-01
3 Volume ratio 1.41 Variation (σ/μ) 7.57e-02 Residual 6.58e-02
4 Volume ratio 1.29 Variation (σ/μ) 4.77e-02 Residual 3.49e-02
5 Volume ratio 1.20 Variation (σ/μ) 3.37e-02 Residual 1.73e-02
6 Volume ratio 1.17 Variation (σ/μ) 2.81e-02 Residual 7.75e-03
7 Volume ratio 1.15 Variation (σ/μ) 2.64e-02 Residual 3.16e-03
8 Volume ratio 1.15 Variation (σ/μ) 2.59e-02 Residual 1.16e-03
9 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 3.88e-04
10 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 1.16e-04
11 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 1.56e-05
12 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 7.57e-06
13 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 3.58e-06
14 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 1.51e-06
15 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 5.71e-07
16 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 1.94e-07
17 Volume ratio 1.15 Variation (σ/μ) 2.57e-02 Residual 5.86e-08
Solver converged in 17 iterations.
Time taken: 2.52 seconds
Plotting the resulting mesh:
from firedrake.pyplot import triplot
fig, axes = plt.subplots()
triplot(mover.mesh, axes=axes)
axes.set_aspect(1)
plt.savefig("monge_ampere_helmholtz-adapted_mesh.jpg")
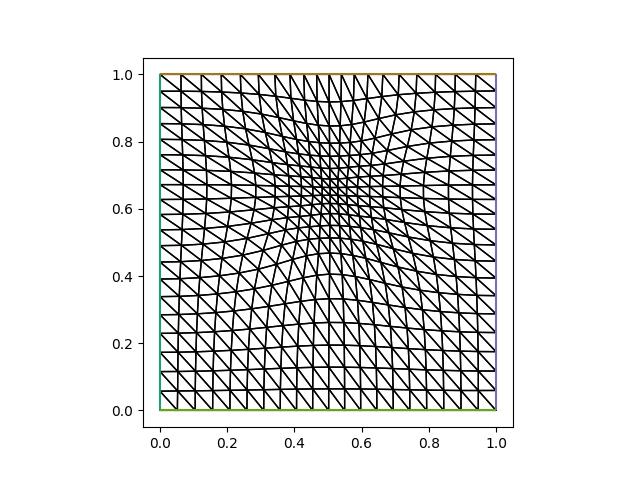
Now let us see whether the numerical error has actually been reduced if we solve the PDE on this mesh:
u_h = solve_helmholtz(mover.mesh)
error = u_h - u_exact(mover.mesh)
print("L2-norm error on moved mesh:", sqrt(assemble(dot(error, error) * dx)))
L2-norm error on moved mesh: 0.005955820168534556
Of course, in many practical problems we do not actually have access to the exact solution. We can then use the Hessian of the numerical solution in the monitor function. When calculating the Hessian we have to be a bit careful however: since our numerical FEM solution \(u_h\) is a piecewise linear function, its first gradient results in a piecewise constant function, a vector-valued constant function in each cell. Taking its gradient in each cell would therefore simply be zero. Instead, we should numerically approximate the derivatives to “recover” the Hessian, for which there are a number of different methods.
As Hessians are often used in metric-based h-adaptivity, some of these
methods have been implemented in the animate package.
An implementation of a monitor based on the Hessian of the numerical
solution is given below. Note that this requires solving the Helmholtz
PDE in every mesh movement iteration, and thus may become significantly
slower for large problems.
from animate import RiemannianMetric
def monitor_solve(mesh):
u_h = solve_helmholtz(mesh)
V = FunctionSpace(mesh, "CG", 1)
TV = TensorFunctionSpace(mesh, "CG", 1)
H = RiemannianMetric(TV)
H.compute_hessian(u_h, method="L2")
Hnorm = Function(V, name="Hnorm")
Hnorm.interpolate(sqrt(inner(H, H)))
Hnorm_max = Hnorm.dat.data.max()
m = 1 + alpha * Hnorm / Hnorm_max
return m
mover = MongeAmpereMover(mesh, monitor_solve, method="quasi_newton", rtol=rtol)
t0 = time.time()
mover.move()
print(f"Time taken: {time.time() - t0:.2f} seconds")
0 Volume ratio 5.04 Variation (σ/μ) 4.90e-01 Residual 4.93e-01
1 Volume ratio 2.60 Variation (σ/μ) 2.27e-01 Residual 2.24e-01
2 Volume ratio 1.63 Variation (σ/μ) 1.09e-01 Residual 1.03e-01
3 Volume ratio 1.31 Variation (σ/μ) 5.30e-02 Residual 4.33e-02
4 Volume ratio 1.20 Variation (σ/μ) 3.31e-02 Residual 1.96e-02
5 Volume ratio 1.16 Variation (σ/μ) 2.65e-02 Residual 8.77e-03
6 Volume ratio 1.14 Variation (σ/μ) 2.46e-02 Residual 3.83e-03
7 Volume ratio 1.14 Variation (σ/μ) 2.40e-02 Residual 1.63e-03
8 Volume ratio 1.14 Variation (σ/μ) 2.38e-02 Residual 6.77e-04
9 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 2.72e-04
10 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 1.06e-04
11 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 3.97e-05
12 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 1.44e-05
13 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 5.30e-06
14 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 2.20e-06
15 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 1.11e-06
16 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 6.13e-07
17 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 3.38e-07
18 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 1.80e-07
19 Volume ratio 1.14 Variation (σ/μ) 2.37e-02 Residual 9.21e-08
Solver converged in 19 iterations.
Time taken: 6.17 seconds
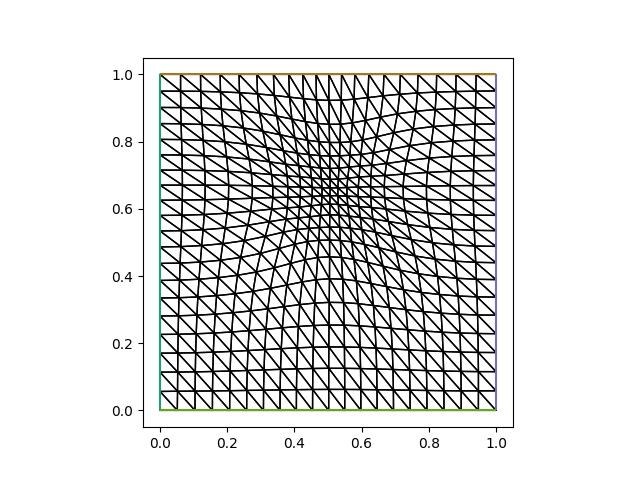
fig, axes = plt.subplots()
triplot(mover.mesh, axes=axes)
axes.set_aspect(1)
plt.savefig("monge_ampere_helmholtz-adapted_mesh2.jpg")
u_h = solve_helmholtz(mover.mesh)
error = u_h - u_exact(mover.mesh)
print("L2-norm error on moved mesh:", sqrt(assemble(dot(error, error) * dx)))
L2-norm error on moved mesh: 0.00630874419681285
As expected, we do not observe significant changes in final adapted meshes between the two approaches. However, the second approach is almost twice longer, as it requires solving the PDE in every iteration. In practice it might therefore be sufficient to compute the solution on the initial mesh and interpolate it onto adapted meshes at every iteration. This is demonstrated in the following implementation:
t0 = time.time()
u_h = solve_helmholtz(mesh)
def monitor_interp_soln(mesh):
V = FunctionSpace(mesh, "CG", 1)
u_h_interp = Function(V).interpolate(u_h)
TV = TensorFunctionSpace(mesh, "CG", 1)
H = RiemannianMetric(TV)
H.compute_hessian(u_h_interp, method="L2")
Hnorm = Function(V, name="Hnorm")
Hnorm.interpolate(sqrt(inner(H, H)))
Hnorm_max = Hnorm.dat.data.max()
m = 1 + alpha * Hnorm / Hnorm_max
return m
mover = MongeAmpereMover(mesh, monitor_interp_soln, method="quasi_newton", rtol=rtol)
mover.move()
print(f"Time taken: {time.time() - t0:.2f} seconds")
0 Volume ratio 5.04 Variation (σ/μ) 4.90e-01 Residual 4.93e-01
1 Volume ratio 1.98 Variation (σ/μ) 8.07e-02 Residual 7.72e-02
2 Volume ratio 1.38 Variation (σ/μ) 4.87e-02 Residual 4.72e-02
3 Volume ratio 1.23 Variation (σ/μ) 3.02e-02 Residual 2.78e-02
4 Volume ratio 1.19 Variation (σ/μ) 1.99e-02 Residual 1.31e-02
5 Volume ratio 1.18 Variation (σ/μ) 1.76e-02 Residual 5.84e-03
6 Volume ratio 1.18 Variation (σ/μ) 1.76e-02 Residual 2.66e-03
7 Volume ratio 1.18 Variation (σ/μ) 1.77e-02 Residual 1.26e-03
8 Volume ratio 1.18 Variation (σ/μ) 1.78e-02 Residual 6.20e-04
9 Volume ratio 1.18 Variation (σ/μ) 1.79e-02 Residual 3.16e-04
10 Volume ratio 1.18 Variation (σ/μ) 1.79e-02 Residual 1.67e-04
11 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 9.05e-05
12 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 5.07e-05
13 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 2.93e-05
14 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 1.74e-05
15 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 1.06e-05
16 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 6.56e-06
17 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 4.13e-06
18 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 2.63e-06
19 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 1.68e-06
20 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 1.08e-06
21 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 6.98e-07
22 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 4.51e-07
23 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 2.91e-07
24 Volume ratio 1.18 Variation (σ/μ) 1.80e-02 Residual 1.88e-07
Solver converged in 24 iterations.
Time taken: 6.19 seconds
fig, axes = plt.subplots()
triplot(mover.mesh, axes=axes)
axes.set_aspect(1)
plt.savefig("monge_ampere_helmholtz-adapted_mesh3.jpg")
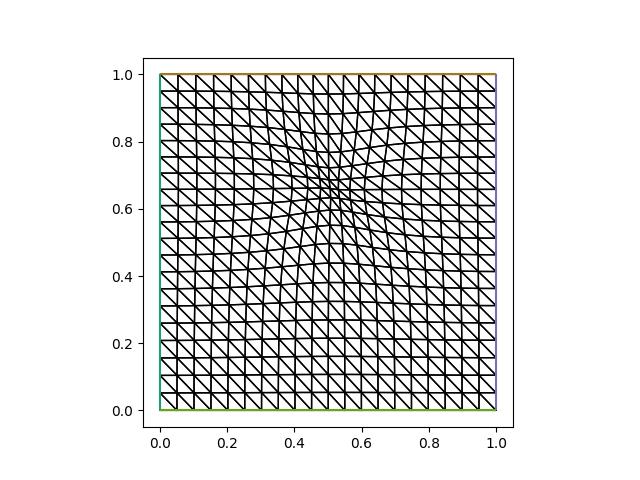
u_h = solve_helmholtz(mover.mesh)
error = u_h - u_exact(mover.mesh)
print("L2-norm error on moved mesh:", sqrt(assemble(dot(error, error) * dx)))
L2-norm error on moved mesh: 0.008712487462902048
Even though the number of iterations has increased (24 compared to 17 and 19), each iteration is slightly cheaper since the PDE is not solved again every time. Despite that, the increased total number of iterations lead to a slightly longer total runtime. Depending on the PDE in question and the solver used to solve it, interpolating the solution may turn out to be slower than recomputing it. We also observe that interpolating the solution lead to a significantly larger error. In conclusion, in this particular example interpolating the solution field did not lead to neither a lower error nor a faster mesh movement step compared to previous approaches. Let us explore one more approach.
We may further speed up the mesh movement step by also avoiding recomputing the Hessian at every iteration, which, in this particular example, is the most expensive part of the monitor function. Similarly to the above example, we compute the solution on the initial (uniform) mesh, but now we also compute its Hessian and interpolate the Hessian onto adapted meshes.
t0 = time.time()
u_h = solve_helmholtz(mesh)
TV = TensorFunctionSpace(mesh, "CG", 1)
H = RiemannianMetric(TV)
H.compute_hessian(u_h, method="L2")
def monitor_interp_Hessian(mesh):
V = FunctionSpace(mesh, "CG", 1)
TV = TensorFunctionSpace(mesh, "CG", 1)
H_interp = RiemannianMetric(TV).interpolate(H)
Hnorm = Function(V, name="Hnorm")
Hnorm.interpolate(sqrt(inner(H_interp, H_interp)))
Hnorm_max = Hnorm.dat.data.max()
m = 1 + alpha * Hnorm / Hnorm_max
return m
mover = MongeAmpereMover(mesh, monitor_interp_Hessian, method="quasi_newton", rtol=rtol)
mover.move()
print(f"Time taken: {time.time() - t0:.2f} seconds")
0 Volume ratio 5.04 Variation (σ/μ) 4.90e-01 Residual 4.93e-01
1 Volume ratio 1.92 Variation (σ/μ) 9.68e-02 Residual 9.18e-02
2 Volume ratio 1.21 Variation (σ/μ) 2.31e-02 Residual 6.69e-03
3 Volume ratio 1.15 Variation (σ/μ) 2.11e-02 Residual 4.14e-05
4 Volume ratio 1.15 Variation (σ/μ) 2.12e-02 Residual 4.80e-08
Solver converged in 4 iterations.
Time taken: 1.09 seconds
fig, axes = plt.subplots()
triplot(mover.mesh, axes=axes)
axes.set_aspect(1)
plt.savefig("monge_ampere_helmholtz-adapted_mesh4.jpg")
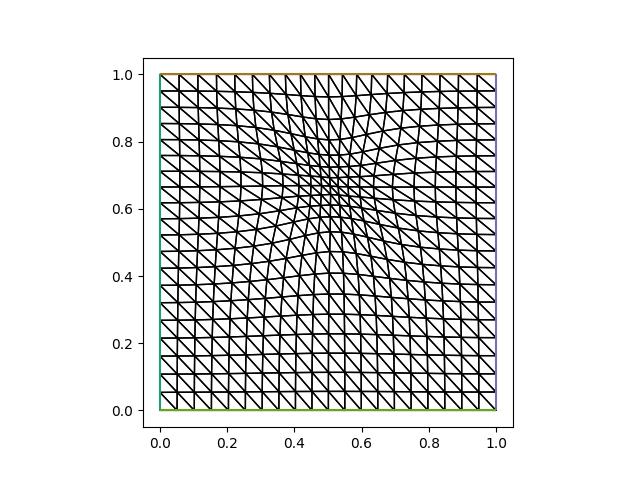
u_h = solve_helmholtz(mover.mesh)
error = u_h - u_exact(mover.mesh)
print("L2-norm error on moved mesh:", sqrt(assemble(dot(error, error) * dx)))
L2-norm error on moved mesh: 0.008385305585746483
The mesh movement step now only took 4 iterations, with a total runtime of only 1.09 seconds, which is up to five times shorter than previous examples. The final error is again larger than the example where the solution is recomputed at every iteration, but is smaller than the example where we interpolated the solution field.
We can summarise these results in the following table:
Monitor function |
Error |
CPU time (s) |
|---|---|---|
|
0.00596 |
2.52 |
|
0.00631 |
6.17 |
|
0.00871 |
6.19 |
|
0.00839 |
1.09 |
In this demo we demonstrated several examples of monitor functions and briefly evaluated their performance. Each approach has inherent advantages and limitations which should be considered for every new problem of interest. PDEs that are highly sensitive to changes in local resolution may require recomputing the solution at every iteration. In other cases we may obtain adequate results by defining a monitor function that computes the solution less frequently, or even only once per iteration. Movement allows and encourages such experimentation with different monitor functions.
This tutorial can be dowloaded as a Python script.
Footnotes